about the group
The research interests in our group are a combination of topics that span the domains of biology, chemistry, data informatics, and high performance computing. We develop and use data analysis techniques, theoretical methods along with large-scale computer simulations to investigate biomolecules. We closely work with experimental collaborators for development and validation of our computational models. We are particularly interested in solving problems that have implications for clean energy, environment and human health.
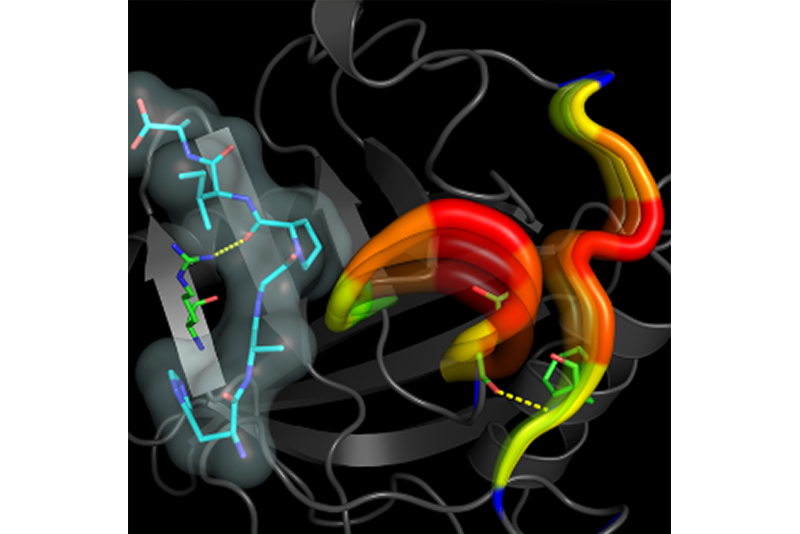
In the area of biology and chemistry, at the molecular level, we are interested in allosteric modulator design for development of safer medicines, and enzyme design/engineering for industrial applications. Using a combination of computational techniques developed in the group over the last decade, we have developed a biophysical model of how enzyme catalysis works. This model provides new insights into how the solvent-enzyme coupling drives enzyme catalysis through long-range interactions in the enzyme structure. This biophysical model is being applied (in collaboration with industrial partners) to development of new allosteric modulators and hypercatalytic enzymes.
In the area of data and computational sciences, we design new big data analysis techniques, computational algorithms and software performance optimization on emerging hardware architectures (including heterogeneous architectures with FPGAs and GPUs). Working with collaborators, we have developed methods to analyze multi-terabytes date sets of high dimensional data for extracting functional features. For productivity optimization, We have optimized and ported simulations code on GPUs with 20 fold speed-up compared to the CPU-only code. The insights gained are being applied to optimize other software codes and methods on the current and future heterogeneous architectures. We are also investigating fault-tolerance and performance auto-tuning strategies for improved end-user productivity on the future computing platforms.
The also develop and distribute open source software for scientific computing. Software developed in the group include VigyaanCD, an open source software workbench for bio/chemical modeling.